
RAFAT Seyed Abbas
- Faculty of Agriculture, University of Tabriz, Tabriz, Iran, Islamic Republic of
- Animal genetics, Genomics, Genomic selection, Small ruminants, Statistical genetics, Structural and functional genomics
- recommender, manager
Recommendations: 3
Reviews: 0
Recommendations: 3

A pipeline with pre-processing options to detect behaviour from accelerometer data using Machine Learning tested on dairy goats.
Introducing a feature extraction method and implementing a gradient boosting algorithm to predict behavioral traits in animals
Recommended by Seyed Abbas Rafat based on reviews by 2 anonymous reviewersIn the livestock industry, the concerns of society and livestock farmers have changed in recent decades, moving from a focus solely on production traits to sustainable production while respecting animal welfare (Ducrot et al., 2024). Animal behavior parameters can serve as a reliable proxy for animal welfare, but collecting large data sets of behavioral data on farm is very time-consuming when not automatized. Accelerometers are promising devices to detect animal behavior. A key element of the efficiency of the prediction of animal behavior from accelerometer data is the adequacy of pre-processing methods (Riaboff et al., 2019,Riaboff et al., 2022, Vidal et al., 2023).
The article of Mauny et al., (2025) aims to find a solution for using huge automatized data. So, livestock farmers can effectively use new technologies to monitor animal behavior and then correct poor husbandry routines. Authors used the pipeline ACT4Behav - (Accelerometer-based Classification Tool for identifying Behaviours) - (Mauny et al., 2024) for the pre-processing of accelerometer data with the aim of selecting the most important features of dairy goats related to rumination, head in the feeder, lying and standing (using data based on video recordings for the validation of behavior activity). Mauny et al., (2025) established a clear methodology that systematically obtain the best features and processing techniques to predict each targeted variable from raw accelerometer data. The work provides valuable information for both animal husbandry specialists and data mining scientists. The main limitation of the work is the small number of animals for both training and model testing.
References
Ducrot C, Barrio MB, Boissy A, Charrier F, Even S, Mormède P, Petit S, Pinard-van der laan M-H, Schelcher F, Casabianca F, Ducos A, Foucras G, Guatteo R, Peyraud J-L, Vayssier-Taussat M, Veysset P, Friggens NC and Fernandez X. 2024. Animal board invited review: Improving animal health and welfare in the transition of livestock farming systems: Towards social acceptability and sustainability. animal 18, 101100. https://doi.org/10.1016/j.animal.2024.101100
Mauny, S, Kwon J, Friggens NC, Duvaux-Ponter C, Taghipoor M. 2024. ACT4Behav (Accelerometer-based Classification Tool for identifying Behaviours): a Machine Learning pipeline with extensive pre-processing and feature creation options. Zenodo. https://doi.org/10.5281/zenodo.12624796
Mauny S, Kwon J, Friggens NC, Duvaux-Ponter C, Taghipoor M. 2025. A pipeline with pre-processing options to detect behaviour from accelerometer data using Machine Learning tested on dairy goats.. Zenodo, ver. 6 peer-reviewed and recommended by PCI Animal Science. https://doi.org/10.5281/zenodo.12627197
Riaboff L, Aubin S, Bédère N, Couvreur S, Madouasse A, Goumand E, Chauvin A, Plantier G. 2019. Evaluation of pre-processing methods for the prediction of cattle behaviour from accelerometer data. Computers and Electronics in Agriculture 165, 104961. https://doi.org/10.1016/j.compag.2019.104961
Riaboff L, Shalloo L, Smeaton AF, Couvreur S, Madouasse A, Keane MT. 2022. Predicting livestock behaviour using accelerometers: A systematic review of processing techniques for ruminant behaviour prediction from raw accelerometer data. Computers and Electronics in Agriculture 192, 106610. https://doi.org/10.1016/j.compag.2021.106610
Vidal G, Sharpnack J, Pinedo P, Tsai IC, Lee AR, Martínez-López B. 2023. Impact of sensor data pre-processing strategies and selection of machine learning algorithm on the prediction of metritis events in dairy cattle. Preventive Veterinary Medicine 215, 105903. https://doi.org/10.1016/j.prevetmed.2023.105903
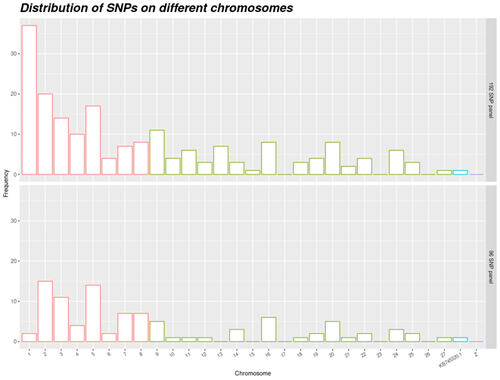
Cost-efficient assignment panel for ducks. Setup of a cost-efficient assignment panel for duck populations.
Providing innovative genetic solutions to the future challenges of the poultry industry: extraction of a small-sized Single-nucleotide polymorphism (SNP) panel using factorial design for parentage assignment in a population consisting of pure and hybrid ducks
Recommended by Seyed Abbas Rafat based on reviews by Arash Javanmard and 2 anonymous reviewersOne of the achievements of animal genetics is that it finds solutions along with the emergence of new needs of the animal husbandry community or the adoption of new laws. Muir and Cheng (2013) research serves as a classic example of using the innovations of animal genetics to meet new legal challenges, such as the restriction of beak cutting in laying hens. Muir and Cheng (2013) investigated the genetic diversity to deal with the cannibalism of intact chickens. In 2021, the European Citizens' Initiative urged the European Commission to legislate against the use of cages for farm animals in the livestock industry. Chapuis et al., (2024) presented a successful solution to the poultry industry about this (future) law by presenting a cost-efficient assignment SNP panel.
In animal breeding, access to pedigree information is necessary for genetic progress. Since the 1970s, the development of genomic science and molecular techniques has shown their ability in this field. Despite the substantial reduction of genotyping costs in the last 20 years, the practical use of genome-wide genotyping for thousands of SNPs remains challenging. Therefore, the search for a small, cost-effective SNP panel is ongoing, with objectives including genetic diversity (Viale et al., 2017), product traceability (Dominik et al., 2021), species and hybrid identification (Harmoinen et al., 2021) and pedigree construction in wild populations (Ekblom et al., 2021). Furthermore, especially in recent decades, small panels of markers have been proposed for parentage assignment in different animals. For example, Domínguez-Viveros et al., (2020) developed panels with 42 to 63 markers for different sheep breeds in Mexico. Similar panels for parentage assignment have been proposed for salmon (May et al., 2020), rainbow trout (Liu et al., 2016), French sheep (Tortereau et al., 2017), Spanish sheep (Calvo et al., 2021), and European bison (Wehrenberg et al., 2024), with marker numbers of 142, 95, 180, 173, and 96, respectively. Massault et al., (2021) showed by simulation that a panel with at least 50 markers is sufficient for progeny assignment in pearl oysters. These examples highlight that extracting a small panel of markers (usually less than 200) from the total genotyping introduced in different species, can open new horizons for applying genomic information in animal breeding.
Chapuis et al. (2024) addressed the challenge of finding an efficient set of markers that can be used in the hybridization of two species of the Pekin duck and the Muscovy duck. They used KASPar technology to setup a panel, with SNPs existing in both species and their hybrids. This panel has sufficient polymorphism to use in practice. Thus, it can be considered as a step forward compared to previous work done on microsatellites. A final list of SNPs was constructed from a reference set comprising 600 K genotyping of Anas platyrhynchos, Cairina moschata and mule duck.
In addition to developing of a cost-efficient assignment panel, the work of Chapuis et al. (2024) presented a factorial design to maintain genetic diversity while considering specificities of duck production. The use of factorial design in avian pedigreed populations is relatively novel, making this research particularly innovative. The study's approach to factorial design in populations with limited size may be generalized to similar poultry species. Furthermore, sufficient effective size of population is selected. So, the panel can be used in other populations outside the tested populations. A notable feature of this panel is the use of neutral SNPs, which ensures that markers will not be lost due to future selection pressures over time.
The paper of Chapuis et al. (2024) exemplifies the application of molecular genetics to address challenges in the poultry industry. The use of kinship matrix instead of relationship matrix, taking into account the unique characteristics of duck production, could be another novelty of the paper. According to the reviewers' comments, the results can be beneficial in the future, particularly with the introduction of the specific factorial design.
References
Calvo JH, Serrano M, Tortereau F, Sarto P, Iguacel LP, Jiménez MA, Folch J, Alabart JL, Fabre S and Lahoz B (2021). Development of a SNP parentage assignment panel in some North-Eastern Spanish meat sheep breeds. Spanish Journal of Agricultural Research 18, e0406. https://doi.org/10.5424/sjar/2020184-16805
Chapuis H, Brard-Fudulea S, Hazard A, Vignal A, Demars J, Rouger R, Teissier M, Gilbert H (2024). Cost-efficient assignment panel for ducks. Setup of a cost-efficient assignment panel for duck populations.: An illustration with experimental data. HAL, hal-04542880, ver. 2 peer-reviewed and recommended by Peer Community in Animal Science. https://hal.inrae.fr/hal-04542880
Domínguez-Viveros J, Rodríguez-Almeida FA, Jahuey-Martínez FJ, Martínez-Quintana JA, Aguilar-Palma GN, Ordoñez-Baquera P (2020). Definition of a SNP panel for paternity testing in ten sheep populations in Mexico, Small Ruminant Research ,193,106262. https://doi.org/10.1016/j.smallrumres.2020.106262
Dominik S, Duff CJ, Byrne AI, Daetwyler H, Reverter A (2021). Ultra-small SNP panels to uniquely identify individuals in thousands of samples. Animal Production Science 61, 1796–1800. https://doi.org/10.1071/AN21123
Ekblom R, Aronsson M, Elsner-Gearing F, Johansson M, Fountain T, Persson J (2021). Sample identification and pedigree reconstruction in Wolverine (Gulo gulo) using SNP genotyping of non-invasive samples. Conservation Genetics Resources 13, 261–274. https://doi.org/10.1007/s12686-021-01208-5
Harmoinen J, von Thaden A, Aspi J, Kvist L, Cocchiararo B, Jarausch A, Gazzola A, Sin T, Lohi H, Hytönen MK, Kojola I, Stronen AV, Caniglia R, Mattucci F, Galaverni M, Godinho R, Ruiz-González A, Randi E, Muñoz-Fuentes V, Nowak C (2021). Reliable wolf-dog hybrid detection in Europe using a reduced SNP panel developed for non-invasively collected samples. BMC Genomics 22, 473. https://doi.org/10.1186/s12864-021-07761-5
Liu S, Palti Y, Gao G, Rexroad CE (2016). Development and validation of a SNP panel for parentage assignment in rainbow trout. Aquaculture 452, 178–182. https://doi.org/10.1016/j.aquaculture.2015.11.001
Massault C, Jones DB, Zenger KR, Strugnell JM, Barnard R, Jerry DR (2021). A SNP parentage assignment panel for the silver lipped pearl oyster (Pinctada maxima). Aquaculture Reports 20, 100687. https://doi.org/10.1016/j.aqrep.2021.100687
May SA, McKinney GJ, Hilborn R, Hauser L, Naish KA (2020). Power of a dual-use SNP panel for pedigree reconstruction and population assignment. Ecology and Evolution 10, 9522–9531. https://doi.org/10.1002/ece3.6645
Muir WM, Cheng HW(2013). Genetics and the Behaviour of Chickens: Welfare and Productivity. In Genetics and the Behaviour of Domestic Animals. Vol. 2 (2nd ed.). pp. 1–30.ISBN: 9780128100165
Tortereau F, Moreno CR, Tosser-Klopp G, Servin B, Raoul J (2017). Development of a SNP panel dedicated to parentage assignment in French sheep populations. BMC Genetics 18, 50. https://doi.org/10.1186/s12863-017-0518-2
Viale E, Zanetti E, Özdemir D, Broccanello C, Dalmasso A, De Marchi M, Cassandro M (2017). Development and validation of a novel SNP panel for the genetic characterization of Italian chicken breeds by next-generation sequencing discovery and array genotyping. Poultry Science 96, 3858–3866. https://doi.org/10.3382/ps/pex238
Wehrenberg G, Tokarska M, Cocchiararo B, Nowak C (2024). A reduced SNP panel optimised for non-invasive genetic assessment of a genetically impoverished conservation icon, the European bison. Scientific Reports 14, 1875. https://doi.org/10.1038/s41598-024-51495-9
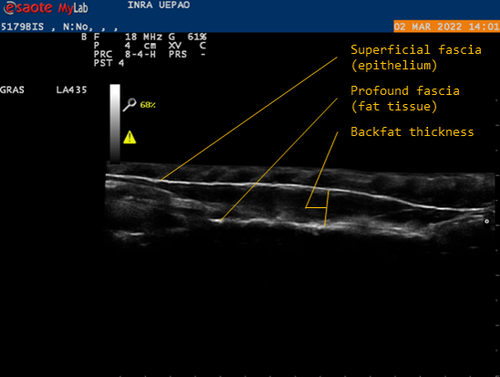
Genetic background of body reserves in laying hens through backfat thickness phenotyping
Towards a better optimization of the genetic improvement of chicken breeds: Introduction of simple phenotypic traits related to body composition for easy measurement in the selection programs of laying hens.
Recommended by Seyed Abbas Rafat based on reviews by 2 anonymous reviewersIn genetic selection, simplistic model of single-trait selection is usually considered, and the response to such approach is estimated using simple models. In practice, however, plant and animal breeders always deal with the selection of several traits, hence making the selection process very complex. Therefore, the simultaneous genetic improvement of several traits has always been one of the goals of livestock, including poultry breeding (Falconer, 1972). Studies that examine the indirect effects of selection on economic traits are eagerly awaited. In this context, the results of the study by Bédère et al., (2023) gives new insights about phenotypic and genotypic relationships between body reserves traits in laying hens. The authors aimed to propose novel data about the genetic architecture of traits related to body fat by measuring a series of phenotypic traits with relatively an easy approach. The authors further aimed to test and validate the phenotyping of backfat thickness as an indicator of the overall fatness of laying hens. Thus, the study allowed providing new evidence regarding the genetic determination of the backfat trait in chicken breeds.
The authors first estimated the effect of selection on the residual feed intake (trait x) on the trait of body reserves (trait y). In fact, divergent selection experiments are a fundamental research tool that allow revealing significant amount of data related to the possible span of genetic improvement for traits of interest. Consequently, by analyzing data from a divergent selection experiment, associations have been estimated between a number of feed-dependent traits that have practical use for chicken breeders. Estimation of the correlations between traits is under question in terms of the theory of genetics and their application in multi-trait selection. As a major finding of the study, the observation of a bimodal distribution of backfat in both lines and the heterogeneity of the variances between families allowed suggesting a possible major gene, which could be investigated in future studies using for instance quantitative genetics. Body composition is continually studied in broilers chicken, but this aspect of chicken genetic is more detailed in laying hens.
The current findings are worthy to validate using several approaches. In fact, one of the limitations of the study can be related to other statistical models that can be built. For example, the study revealed high correlations between egg production and body weight, thus body weight could be considered as a covariate in regression models. Moreover, the principal trait of selection (based on the residual feed intake) could be considered.
References:
Falconer, D. S. (1972). Introduction to Quantitative Genetics. Publisher: Ronald Press Company. pp 365.
Bédère, N., Dupont, J., Baumard, Y., Staub, C., Gourichon, D., Elleboudt, F., Le Roy, P., Zerjal, T. (2023). Genetic background of body reserves in laying hens through backfat thickness phenotyping. HAL ver. 3 peer-reviewed and recommended by Peer Community in Animal Science. https://hal.inrae.fr/hal-04172576