Latest recommendations

| Id | Title | Authors | Abstract | Picture▼ | Thematic fields | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
11 Dec 2023
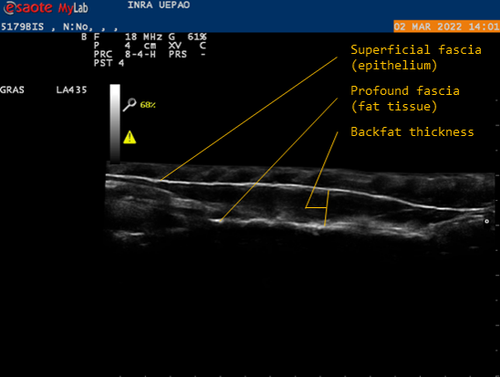
Genetic background of body reserves in laying hens through backfat thickness phenotypingNicolas Bédère, Joëlle Dupont, Yannick Baumard, Christophe Staub, David Gourichon, Frédéric Elleboudt, Pascale Le Roy, Tatiana Zerjal https://hal.inrae.fr/hal-04172576Towards a better optimization of the genetic improvement of chicken breeds: Introduction of simple phenotypic traits related to body composition for easy measurement in the selection programs of laying hens.Recommended by Seyed Abbas Rafat based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
In genetic selection, simplistic model of single-trait selection is usually considered, and the response to such approach is estimated using simple models. In practice, however, plant and animal breeders always deal with the selection of several traits, hence making the selection process very complex. Therefore, the simultaneous genetic improvement of several traits has always been one of the goals of livestock, including poultry breeding (Falconer, 1972). Studies that examine the indirect effects of selection on economic traits are eagerly awaited. In this context, the results of the study by Bédère et al., (2023) gives new insights about phenotypic and genotypic relationships between body reserves traits in laying hens. The authors aimed to propose novel data about the genetic architecture of traits related to body fat by measuring a series of phenotypic traits with relatively an easy approach. The authors further aimed to test and validate the phenotyping of backfat thickness as an indicator of the overall fatness of laying hens. Thus, the study allowed providing new evidence regarding the genetic determination of the backfat trait in chicken breeds. The authors first estimated the effect of selection on the residual feed intake (trait x) on the trait of body reserves (trait y). In fact, divergent selection experiments are a fundamental research tool that allow revealing significant amount of data related to the possible span of genetic improvement for traits of interest. Consequently, by analyzing data from a divergent selection experiment, associations have been estimated between a number of feed-dependent traits that have practical use for chicken breeders. Estimation of the correlations between traits is under question in terms of the theory of genetics and their application in multi-trait selection. As a major finding of the study, the observation of a bimodal distribution of backfat in both lines and the heterogeneity of the variances between families allowed suggesting a possible major gene, which could be investigated in future studies using for instance quantitative genetics. Body composition is continually studied in broilers chicken, but this aspect of chicken genetic is more detailed in laying hens. The current findings are worthy to validate using several approaches. In fact, one of the limitations of the study can be related to other statistical models that can be built. For example, the study revealed high correlations between egg production and body weight, thus body weight could be considered as a covariate in regression models. Moreover, the principal trait of selection (based on the residual feed intake) could be considered. References: Falconer, D. S. (1972). Introduction to Quantitative Genetics. Publisher: Ronald Press Company. pp 365. Bédère, N., Dupont, J., Baumard, Y., Staub, C., Gourichon, D., Elleboudt, F., Le Roy, P., Zerjal, T. (2023). Genetic background of body reserves in laying hens through backfat thickness phenotyping. HAL ver. 3 peer-reviewed and recommended by Peer Community in Animal Science. https://hal.inrae.fr/hal-04172576 | Genetic background of body reserves in laying hens through backfat thickness phenotyping | Nicolas Bédère, Joëlle Dupont, Yannick Baumard, Christophe Staub, David Gourichon, Frédéric Elleboudt, Pascale Le Roy, Tatiana Zerjal | <p>In this study, we pursued three primary objectives: firstly to test and validate the phenotyping of backfat thickness as an indicator of the overall fatness of laying hens; secondly, to estimate genetic parameters for this trait; thirdly, to st... |  | Animal genetics, Poultry, Statistical genetics | Seyed Abbas Rafat | 2023-07-27 17:09:10 | View | |
14 Dec 2022
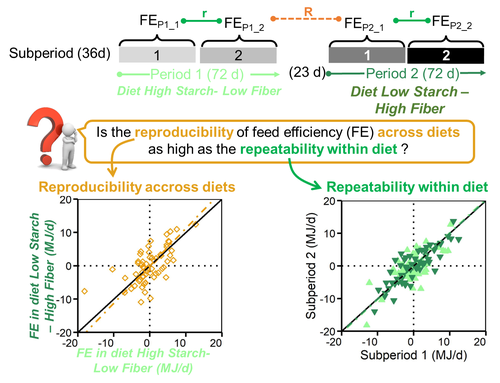
Feed efficiency of lactating Holstein cows was not as repeatable across diets as within diet over subsequent lactation stagesAmelie Fischer, Philippe Gasnier, philippe faverdin https://doi.org/10.1101/2021.02.10.430560A focus on feed efficiency reproducibility and repeatability of dairy cows fed different diets over the lactation stage.Recommended by Alberto Atzori based on reviews by Ioannis Kaimakamis, Angela Schwarm and 2 anonymous reviewersThe topic of feed efficiency is under discussion in the scientific community and several studies pointed out that lactation stage has to be accounted for when estimates of feed efficiency are carried out, especially for genetic ranking of animals and their performances, as highlighted by Li et al. (2017). Other researchers applied a latin square design to test dietary effects across lactation (Ipharraguerre et al. 2002) but this approach cannot be followed out of experimental conditions and particularly does not allow, nowadays, to valorize precision livestock farm data to get phenotypic information from individual animals at farm level. The current manuscript by Fischer, et al. (2022a) describes an experimental trial in which cows were first fed a high starch diet-low fibre then switched over to a low starch diet-high fibre and individually monitored over time. Data were analyzed with the objective to investigate effects within diets and across diets. Since all cows went through the same sequence at the same time it was not possible to completely separate the confounding effect of lactation stage and diet as stated by the authors. However, this manuscript adds methodological discussions and opens research questions especially to the matter of repeatability and reproducibility of feed efficiency of individual animals over the lactation stage. These variables are fundamental to evaluate nutritional traits and phenotypic performances of dairy cows at farm level, as highlighted by a paper of the same first author (Fischer, et al. 2022b) dealing to reproducibility and repeatability with a similar approach. My opinion is that this manuscript gives the opportunity to enlarge the scientific discussions on the calculation of repeatability and reproducibility of feed efficiency of individual animals over time. In particular, as in this study, specific mathematical approaches need to be carried out with the final goal to analyze and valorize precision livestock farm data for cow phenotyping and to propose new methods of feed efficiency evaluations. It also needs complete databases carried out under experimental conditions. In fact it has to be considered that this manuscript makes available to the scientific community all the data and the R code developed for data analysis giving the opportunity to replicate the calculations and propose new advancements in the feed efficiency evaluations of dairy cows. References Fischer A, Gasnier P, Faverdin P (2022a) Feed efficiency of lactating Holstein cows was not as repeatable across diets as within diet over subsequent lactation stages. bioRxiv, 2021.02.10.430560, ver. 3 peer-reviewed and recommended by Peer Community in Animal Science. https://doi.org/10.1101/2021.02.10.430560 Fischer A, Dai X, Kalscheur KF (2022b) Feed efficiency of lactating Holstein cows is repeatable within diet but less reproducible when changing dietary starch and forage concentrations. animal, 16, 100599. https://doi.org/10.1016/J.ANIMAL.2022.100599 Ipharraguerre IR, Ipharraguerre RR, Clark JH (2002) Performance of Lactating Dairy Cows Fed Varying Amounts of Soyhulls as a Replacement for Corn Grain. Journal of Dairy Science, 85, 2905–2912. https://doi.org/10.3168/JDS.S0022-0302(02)74378-6 Li B, Berglund B, Fikse WF, Lassen J, Lidauer MH, Mäntysaari P, Løvendahl P (2017) Neglect of lactation stage leads to naive assessment of residual feed intake in dairy cattle. Journal of Dairy Science, 100, 9076–9084. https://doi.org/10.3168/JDS.2017-12775
| Feed efficiency of lactating Holstein cows was not as repeatable across diets as within diet over subsequent lactation stages | Amelie Fischer, Philippe Gasnier, philippe faverdin | <p> Background: Improving feed efficiency has become a common target for dairy farmers to<br>meet the requirement of producing more milk with fewer resources. To improve feed<br>efficiency, a prerequisite is to ensure that the cows identified... |  | Cattle production, Ruminant nutrition | Alberto Atzori | Anonymous, Ioannis Kaimakamis, Giuseppe Conte, Angela Schwarm | 2021-02-11 08:43:59 | View |
29 Jan 2024
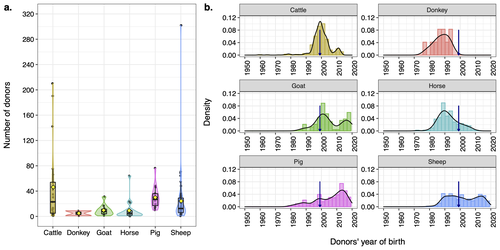
Assessing the potential of germplasm collections for the management of genetic diversity: the case of the French National CryobankAlicia Jacques, Delphine Duclos, Coralie Danchin-Burge, Marie-Jose Mercat, Michele Tixier-Boichard, Gwendal Restoux https://doi.org/10.1101/2023.07.19.549644Exploring Genetic Diversity Management: Unveiling the Potential of Germplasm Collections in the French National CryobankRecommended by Yuliaxis Ramayo-Caldas based on reviews by Roy Costilla and 1 anonymous reviewerThe study by Jacques et al. (2024) addresses a critical concern in the context of genetic diversity erosion in domesticated animal populations. The research uses data from the cryopreserved resources from the French National Cryobank to manage genetic diversity in livestock species. The authors employ a comprehensive methodology to propose novel biodiversity metrics to characterize the status of genetic diversity of cryopreserved collections including cattle, sheep, goat, horse, donkey, and pig livestock species. The findings reveal significant variations of genetic diversity at species and breed levels. Breeds with a large commercial distribution had more donors in the collection than local breeds. The authors propose a practical framework for assessing germplasm collections, providing a valuable tool for planning and managing collections at both national and international levels. The study also highlights the usefulness of the Gini-Simpson and effective donor numbers indices to plan a more efficient sampling, whereas the index of diversity impact can be employed in the selection of the most suitable donors for immediate use, based on pedigree but also using genetic markers. In resume, this study makes a significant contribution to the field by offering a framework for the assessment of germplasm collections. Its innovative metrics provide insights that could guide strategic decision-making in planning, managing, and utilizing cryopreserved resources. This research is relevant and can benefit conservationists, and population genetics working towards the preservation and sustainable use of genetic resources in livestock species. Reference Jacques, A., Duclos, D., Danchin-Burge, C., Mercat, M. J., Tixier-Boichard M., Restoux, G. (2024). Assessing the potential of germplasm collections for the management of genetic diversity: the case of the French National Cryobank. bioRxiv 2023.07.19.549644. ver. 3 peer-reviewed and recommended by Peer Community in Animal Science. https://doi.org/10.1101/2023.07.19.549644
| Assessing the potential of germplasm collections for the management of genetic diversity: the case of the French National Cryobank | Alicia Jacques, Delphine Duclos, Coralie Danchin-Burge, Marie-Jose Mercat, Michele Tixier-Boichard, Gwendal Restoux | <p>Through a combination of selective pressure and genetic drift, there has been a notable erosion of genetic diversity in domesticated animal populations. In response, many countries, including France, have developed gene banks in order to conser... |  | Animal genetics | Yuliaxis Ramayo-Caldas | 2023-07-20 19:08:40 | View | |
31 Jul 2023

The big challenge for livestock genomics is to make sequence data payMartin Johnsson https://doi.org/10.48550/arXiv.2302.01140The price of sequencing the livestock genomicsRecommended by Marcin Pszczoła based on reviews by Mario Calus and 1 anonymous reviewerUsing sequence data in livestock genomics has often been regarded as a solution to revolutionize livestock breeding (Meuwissen & Goddard, 2010). The main expected benefits were to enhance the accuracy of breeding values, achieve better persistence of the accuracy over generations, and enable across populations or breed predictions (Hickey, 2013). Despite the promised benefits, whole-genome sequencing has not yet been implemented in livestock breeding programs, replacing SNP arrays for routine evaluation. In this work, Johnsson (2023) thoroughly reviewed the literature regarding the implications of whole-genome sequencing and functional genomics for livestock breeding practice. The author discusses the potential applications and reasons for difficulties in their implementation. The author speculates that the main challenge for making using the sequence data profitable is to overcome the problem of the small dimensionality of the genetic data and proposes three potential ways to achieve this goal. The first approach is better modeling of genomic segments, the second inclusion of undetected genetic variation, and the third use of functional genomic information. The paper presents an original and interesting perspective on the current status of the use of sequence data in livestock breeding programs and perspectives for the future. References Hickey,J.M.,2013.Sequencing millions of animals for genomic selection 2.0. Journal of Animal Breeding and Genetics 130:331–332. https://doi.org/10.1111/jbg.12054 Johnsson, M., 2023. The big challenge for livestock genomics is to make sequence data pay. arXiv, 2302.01140, ver. 4 peer-reviewed and recommended by Peer Community in Animal Science. https://doi.org/10.48550/arXiv.2302.01140 Meuwissen, T., Goddard, M.,2010. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185:623–631. https://doi.org/10.1534/genetics.110.116590
| The big challenge for livestock genomics is to make sequence data pay | Martin Johnsson | <p>This paper will argue that one of the biggest challenges for livestock genomics is to make whole-genome sequencing and functional genomics applicable to breeding practice. It discusses potential explanations for why it is so difficult to consis... |  | Genomics, Genomic selection | Marcin Pszczoła | 2023-02-03 08:08:39 | View | |
28 Jan 2022
Microbial colonization of tannin-rich tropical plants: interplay between degradability, methane production and tannin disappearance in the rumenMoufida Rira, Diego P Morgavi, Milka Popova, Gaelle Maxin, Michel Doreau https://doi.org/10.1101/2021.08.12.456105Ruminal microbial degradation of tannin-rich tropical plants and methane productionRecommended by Antonio Faciola based on reviews by Todd Callaway and Srinivasan MahalingamRira et al. (2022) evaluated ruminal degradation of tropical tannins-rich plants and the relationship between condensed tannins disappearance and microbial communities. I found this study relevant because a major limitation for tropical plants utilization by ruminants is their potential reduced nutrient digestion. In this study, authors used leaves from Calliandra calothyrsus, Gliricidia sepium, and Leucaena leucocephala, pods from Acacia nilotica and the leaves of Manihot esculenta and Musa spp., which were incubated in situ in the rumen of dairy cows. An in vitro approach was also used to assess the effects of these plants on ruminal fermentation. They observed that hydrolysable and free condensed tannins from all plants completely disappeared after 24 h incubation in the rumen. Disappearance of protein-bound condensed tannins was variable with values ranging from 93% for Gliricidia sepium to 21% for Acacia nilolitica. This demonstrated some potential for selection and improvements in protein digestion. In contrast, fibre-bound condensed tannins disappearance averaged ~82% and did not vary between plants, which was remarkable. The authors noted that disappearance of bound fractions of condensed tannins was not associated with degradability of plant fractions and that the presence of tannins interfered with the microbial colonisation of plants. Each plant had distinct bacterial and archaeal communities after 3 and 12 h of incubation in the rumen and distinct protozoal communities at 3 h. This suggests a great deal of specificity for microbial-plant interactions, which warrants further evaluation to consider also animal contributions to such specificity. Adherent communities in tannin-rich plants had a lower relative abundance of fibrolytic microbes, notably Fibrobacter spp. Whereas, archaea diversity was reduced in high tannin-containing Calliandra calothyrsus and Acacia nilotica at 12 h of incubation. Concurrently, in vitro methane production was lower for Calliandra calothyrsus, Acacia nilotica and Leucaena leucocephala although for the latter total volatile fatty acids production was not affected and was similar to control. Finally, the study demonstrated that the total amount of hydrolysable and condensed tannins contained in a plant play a role governing the interaction with rumen microbes affecting degradability and fermentation. The effect of protein- and fibre-bound condensed tannins on degradability is less important. The major limitation of the study is the lack of animal validation at this stage; therefore, further studies are warranted, especially studies evaluating these plants in vivo. Furthermore, mechanisms associated with plant-microbial specificity, the role played by the host, and more data on nutrient utilization and gas production should be investigated. Nonetheless, this work show interesting microbial colonization and specific plant-microbial relationships that are novel in the ruminal environment. Reference: Rira M, Morgavi DP, Popova M, Maxin G, Doreau M (2022). Microbial colonization of tannin-rich tropical plants: interplay between degradability, methane production and tannin disappearance in the rumen. bioRxiv, 2021.08.12.456105, ver. 3 peer-reviewed and recommended by Peer Community in Animal Science. https://doi.org/10.1101/2021.08.12.456105
| Microbial colonization of tannin-rich tropical plants: interplay between degradability, methane production and tannin disappearance in the rumen | Moufida Rira, Diego P Morgavi, Milka Popova, Gaelle Maxin, Michel Doreau | <p>Condensed tannins in plants are found free and attached to protein and fibre but it is not<br>known whether these fractions influence rumen degradation and microbial colonization.<br>This study explored the rumen degradation of tropical tannins... |  | Animal nutrition modelling, Cattle production, Emissions , Farming systems, Gut microbiology, Microbial ecology, Microbial fermentation, Rumen microbiology, Rumen microbiome , Ruminant nutrition | Antonio Faciola | 2021-08-16 08:56:45 | View | |
07 Feb 2022

Resilience: reference measures based on longer-term consequences are needed to unlock the potential of precision livestock farming technologies for quantifying this traitFriggens, N. C., Adriaens, I., Boré, R., Cozzi, G., Jurquet, J., Kamphuis, C., Leiber, F., Lora, I., Sakowski, T., Statham, J. and De Haas, Y. https://doi.org/10.5281/zenodo.5215797Measuring resilience in farm animals: theoretical considerations and application to dairy cowsRecommended by Aurélien Madouasse based on reviews by Ian Colditz and 2 anonymous reviewers based on reviews by Ian Colditz and 2 anonymous reviewers
Farm animals differ in their ability to respond to the many environmental challenges they face. Such challenges include infectious diseases, metabolic diseases resulting from inadequate coverage of dietary needs, as well as the diverse consequences of climate change. Various concepts exist to characterise the responses of animals to different types of challenges. This article by Friggens et al. (2022) focuses on resilience, providing a conceptual definition and proposing a method to quantify resilience in dairy cows. The first part of the paper provides a definition of resilience and highlights its differences and relations with the related concepts of robustness, and, to a lesser extent, resistance and tolerance. In essence, resilience is the ability of an animal to bounce back quickly after a challenge of limited duration. On the other hand, robustness is the ability of an animal to cope with conditions that are overall unfavourable. From these conceptual and intuitive definitions, there are several difficulties precluding the design of concrete methods to measure resilience. First, there is some degree of overlap between the concepts of resilience, robustness, resistance and tolerance. Secondly, resilience is a multidimensional concept whereby resilience to a given perturbation does not imply resilience to other types of perturbation, e.g. resilience to a challenge by a specific pathogen does not imply resilience to a nutritional challenge. A further difficulty in the measure of resilience is the fact that different animals may be exposed to challenges that are different in nature and in number. The authors argue that although resilience cannot be measured directly (it should be seen as a latent construct), it is possible to quantify it indirectly through its consequences. In the second part of the paper, the authors propose a method to quantify resilience of individual dairy cows. The method is based on the premise that resilient animals should be kept longer in their herd than non-resilient animals. The main criterion in the evaluation is therefore the ability of cows to re-calve. Each cow that is calving receives a certain number of points, to which, in each lactation, bonus points are added for higher milk production and penalty points are removed for each insemination after the first one, for each disease event and for each day of calving interval above some herd specific value. Therefore, cows have a resilience score in each lactation. They also have a lifetime resilience score obtained by summing the scores for all the lactations, that gets bigger as the cow has more calves, and that also takes the age at first calving into account. In a previous study, Adriaens et al. (2020) showed that higher resilience scores were associated with fewer drops in milk yield and more stable activity dynamics. Starting from theoretical considerations on the notion of resilience, this paper describes a concrete method to quantify animal-level resilience on farm. Such quantification will be useful for breeding and culling decisions. Finally, the general framework to design resilience measures that is presented will be useful to researchers working on the quantification of farm animal resilience using new methods and data sources.
References Adriaens I, Friggens NC, Ouweltjes W, Scott H, Aernouts B and Statham J 2020. Productive life span and resilience rank can be predicted from on-farm first-parity sensor time series but not using a common equation, across farms. Journal of Dairy Science 103, 7155-7171.https://doi.org/10.3168/jds.2019-17826 Friggens, N.C. , Adriaens, I., Boré, R., Cozzi, G., Jurquet, J., Kamphuis, C., Leiber, F., Lora, I., Sakowski, T., Statham, J., De Haas, Y. (2022). Resilience: reference measures based on longer-term consequences are needed to unlock the potential of precision livestock farming technologies for quantifying this trait. Zenodo, 5215797, ver. 5 peer-reviewed and recommended by Peer community in Animal Science. https://dx.doi.org/10.5281/zenodo.5215797 | Resilience: reference measures based on longer-term consequences are needed to unlock the potential of precision livestock farming technologies for quantifying this trait | Friggens, N. C., Adriaens, I., Boré, R., Cozzi, G., Jurquet, J., Kamphuis, C., Leiber, F., Lora, I., Sakowski, T., Statham, J. and De Haas, Y. | <p style="text-align: justify;">Climate change, with its increasing frequency of environmental disturbances puts pressures on the livestock sector. To deal with these pressures, more complex traits such as resilience must be considered in our mana... |  | Precision livestock farming | Aurélien Madouasse | 2021-08-20 15:34:13 | View | |
31 Jan 2020

OneARK: Strengthening the links between animal production science and animal ecologyDelphine Destoumieux-Garzón, Pascal Bonnet, Céline Teplitsky, François Criscuolo, Pierre-Yves Henry, David Mazurais, Patrick Prunet, Gilles Salvat, Philippe Usseglio-Polatera, Etienne Verrier and Nicolas Friggens https://doi.org/10.5281/zenodo.3632731When scientific communities intertwineRecommended by Pauline Ezanno based on reviews by Rowland Raymond Kao, Arata Hidano and 1 anonymous reviewerScientific research can be seen by some as a competitive territory: competition of opinions, concepts, publications, competition for funding. Fortunately, it is above all a territory of sharing and cross-fertilization of ideas. It is gradually becoming a territory of productive interdisciplinary collaborations, despite persistent resistance to making borders more permeable [1]. At the crossroads of worlds, many challenges must be met for communities to understand each other, to be able to communicate with one another, and to benefit mutually from scientific interactions [2]. Delphine Destoumieux-Garzon and co-authors [3] propose to stimulate a single Animal Research Kinship (OneARK) to promote the crossing of the scientific communities in animal production and animal ecology. These two communities share many concepts and methods, which, while they are based on marked specificities (natural versus artificial systems), also and above all have common points that need to be explored more closely. Seven concepts of shared interest to improve the resilience and sustainability of animal population systems were explored by the authors: selection, system viability, system management, animal adaptability, inter-individual diversity in systems, agroecology, and animal monitoring. This foundation stone paves the way for a finer integration between these two communities, which are close and yet distant, and which are slowly getting to know, understand, and recognize each other. References [1] Ledford, H. (2015). How to solve the world’s biggest problems. Nature, 525, 308–311. doi: 10.1038/525308a | OneARK: Strengthening the links between animal production science and animal ecology | Delphine Destoumieux-Garzón, Pascal Bonnet, Céline Teplitsky, François Criscuolo, Pierre-Yves Henry, David Mazurais, Patrick Prunet, Gilles Salvat, Philippe Usseglio-Polatera, Etienne Verrier and Nicolas Friggens | <p>1. Wild and farmed animals are key elements of natural and managed ecosystems that deliver functions such as pollination, pest control and nutrient cycling within the broader roles they play in contributing to biodiversity and to every category... |  | Agricultural sustainability, Animal genetics, Animal welfare, Ecology, Precision livestock farming, Veterinary epidemiology | Pauline Ezanno | 2019-07-05 15:33:21 | View | |
09 Apr 2022

The impact of housing conditions on porcine mesenchymal stromal/stem cell populations differ between adipose tissue and skeletal muscleAudrey Quéméner, Frédéric Dessauge, Marie-Hélène Perruchot, Nathalie Le Floc’h, Isabelle Louveau https://doi.org/10.1101/2021.06.08.447546Housing conditions affect cell populations in adipose and muscle tissues of pigsRecommended by Hervé Acloque based on reviews by 2 anonymous reviewersThe adaptability of livestock to changing environments is based in particular on their genetic characteristics but also on the farming conditions to which they are subjected. However, this last point is poorly documented and little is known about its contribution to environmental challenges. The study by Quéméner and colleagues [1] addresses this question by assessing the effect of two hygiene conditions (good vs poor) on the distribution of cell populations present in adipose and muscle tissues of pigs divergently selected for feed efficiency [2]. The working hypothesis is that degraded housing conditions would be at the origin of an hyper stimulation of the immune system that can influence the homeostasis of adipose tissue and skeletal muscle and consequently modulate the cellular content of these tissues. Cellular compositions are thus interesting intermediate phenotypes for quantifying complex traits. The study uses pigs divergently selected for residual feed intake (RFI+ and RFI-) to assess whether there is a genetic effect associated with the observed phenotypes. The study characterized different stromal cell populations based on the expression of surface markers: CD45 to separate hematopoietic lineages and markers associated with the stem properties of mesenchymal cells: CD56, CD34, CD38 and CD140a. The authors observed that certain subpopulations are differentially enriched according to the hygiene condition (good vs poor) in adipose and skeletal tissue (CD45-CD56-) sometimes with an associated (genetic) lineage effect. This pioneering study validates a number of tools for characterizing cell subpopulations present in porcine adipose and muscle tissue. It confirms that housing conditions can have an effect on intermediate phenotypes such as intra-tissue cell populations. This pioneering work will pave the way to better understand the effects of livestock systems on tissue biology and animal phenotypes and to characterize the nature and function of progenitor cells present in muscle and adipose tissue. [1] Quéméner A, Dessauge F, Perruchot MH, Le Floc’h N, Louveau I. 2022. The impact of housing conditions on porcine mesenchymal stromal/stem cell populations differ between adipose tissue and skeletal muscle. bioRxiv 2021.06.08.447546, ver. 3 peer-reviewed and recommended by Peer Community in Animal Science. https://doi.org/10.1101/2021.06.08.447546 [2] Gilbert H, Bidanel J-P, Gruand J, Caritez J-C, Billon Y, Guillouet P, Lagant H, Noblet J, Sellier P. 2007. Genetic parameters for residual feed intake in growing pigs, with emphasis on genetic relationships with carcass and meat quality traits. Journal of Animal Science 85:3182–3188. https://doi.org/10.2527/jas.2006-590. | The impact of housing conditions on porcine mesenchymal stromal/stem cell populations differ between adipose tissue and skeletal muscle | Audrey Quéméner, Frédéric Dessauge, Marie-Hélène Perruchot, Nathalie Le Floc’h, Isabelle Louveau | <p><strong>Background.</strong> In pigs, the ratio between lean mass and fat mass in the carcass determines production efficiency and is strongly influenced by the number and size of cells in tissues. During growth, the increase in the number of c... |  | Monogastrics, Physiology, Veterinary science | Hervé Acloque | 2021-06-08 17:34:54 | View | |
14 Oct 2020

Determining insulin sensitivity from glucose tolerance tests in Iberian and Landrace pigsJ. M. Rodríguez-López, M. Lachica, L. González-Valero, I. Fernández-Fígares https://doi.org/10.1101/2019.12.20.884056Iberian pigs: more than excellent ham!Recommended by Jordi Estellé based on reviews by 2 anonymous reviewersIberian pigs represent a treasured resource that allows the maintenance of their “montanera” traditional breeding system and, thus, contributes to the socioeconomic sustainability of the rural areas in the south-western regions of Iberian Peninsula. While the excellence of Iberian meat products is widely recognized, the idea of using Iberian pigs as biomedical models is currently emerging. Interestingly, due to the particular fatty acid metabolism of this porcine breed, Iberian pigs have been proposed as models for type 2 diabetes (Torres-Rovira et al. 2012) or obesity-related renal disease (Rodríguez et a. 2020). In the present manuscript, Rodríguez-López et al. provide further insights on the particularities of “obese” Iberian pigs by comparing their insulin sensitivity in a glucose tolerance test with that of commercial “lean” Landrace pigs. The authors compared four Iberian pigs with five Landrace pigs in an intense time-series following an intra-arterial glucose tolerance test and measuring insulin, glucose, lactate, triglycerides, cholesterol, creatinine, albumin and urea plasma levels. Several of these parameters showed significant differences between both breeds, with some of them being compatible with an early stage of insulin resistance in Iberian pigs. These results are relevant from an animal production perspective, but provide also further evidence for considering the Iberian pigs as a suitable biomedical model for obesity-related disorders. References [1] Torres-Rovira, L., Astiz, S., Caro, A., Lopez-Bote, C., Ovilo, C., Pallares, P., Perez-Solana, M. L., Sanchez-Sanchez, R., & Gonzalez-Bulnes, A. (2012). Diet-induced swine model with obesity/leptin resistance for the study of metabolic syndrome and type 2 diabetes. The Scientific World Journal, 510149. https://doi.org/10.1100/2012/510149 | Determining insulin sensitivity from glucose tolerance tests in Iberian and Landrace pigs | J. M. Rodríguez-López, M. Lachica, L. González-Valero, I. Fernández-Fígares | <p>As insulin sensitivity may help to explain divergences in growth and body composition between native and modern breeds, metabolic responses to glucose infusion were measured using an intra-arterial glucose tolerance test (IAGTT). Iberian (n = 4... |  | Monogastrics, Physiology, Pig nutrition | Jordi Estellé | 2019-12-28 10:51:03 | View | |
02 Sep 2021

A modelling framework for the prediction of the herd-level probability of infection from longitudinal dataAurélien Madouasse, Mathilde Mercat, Annika van Roon, David Graham, Maria Guelbenzu, Inge Santman Berends, Gerdien van Schaik, Mirjam Nielen, Jenny Frössling, Estelle Ågren, Roger Humphry, Jude Eze, George Gunn, Madeleine Henry, Jörn Gethmann, Simon J. More, Nils Toft, Christine Fourichon https://doi.org/10.1101/2020.07.10.197426Modelling freedom from disease - how do we compare between countries?Recommended by Rowland Raymond Kao based on reviews by Arata Hidano and 1 anonymous reviewerIn this paper, Madouasse et al. (2021) present a generalisable Bayesian method for calculating the probability that a herd is free from disease, based on its prior disease status, and using data (herd status over time over a sufficient number of herds to inform the model) and reasonable prior estimates of the sensitivity and specificity of tests being used to determine animal infection status. Where available, the modelling approach can also include relevant additional risk factors. By bringing all these factors together, it allows for most countries to use the same analytical approach on their data, with differences across datasets expressed in terms of the uncertainty around the central estimates. Having a single methodology that generates both a central estimate of disease freedom, and uncertainty thus provides the opportunity (given typically available data) to compare the probability of freedom across different systems. This is relevant in terms of the context of trade (since international trade of livestock in many cases depends on disease freedom). It is also important when evaluating, for example, transnational burdens of disease - and with different regulations in place in different countries, this is invaluable and can be used, for example, to assess risks of zoonotic infection including for zoonotic infection emergence. In the BVD example provided, the point is made that, since regular testing would probably pick up infection rapidly, the addition of risk factors is most valuable where testing is infrequent. This emphasizes the advantages of direct incorporation of risk factors into a single modelling framework. From a technical point of view, the analysis compares two different packages for the Markov Chain Monte Carlo (MCMC) implementation necesary to run the model. They show that, while there are some slight systematic differences, the estimates provided by the two methods are similar to each other; as one method is approximate but substantially more stable and generally much more computationally efficient, this is an important outcome. Both implementations are freely available and with relevant additional software made similarly available by the authors. This is extremely welcome and should encourage its general adoption across different countries. No single model can of course account for everything. In particular, the reliance on past data means that there is an implicit assumption common to all purely statistical methods that the underlying risks have not changed. Thus projections to altered circumstances (changing underlying risk factors or systematic changes in testing or test performance) cannot so easily be incorporated, since these factors are complicated by the dynamics of infection that lie outside the modelling approach. Of course the well known quote from George Box that "all models are wrong" applies here - the generality of approach, statistical robustness and open source philosophy adopted make this model very useful indeed. Madouasse A, Mercat M, van Roon A, Graham D, Guelbenzu M, Santman Berends I, van Schaik G, Nielen M, Frössling J, Ågren E, Humphry RW, Eze J, Gunn GJ, Henry MK, Gethmann J, More SJ, Toft N, Fourichon C (2021) A modelling framework for the prediction of the herd-level probability of infection from longitudinal data. bioRxiv, 2020.07.10.197426, ver. 6 peer-reviewed and recommended by PCI Animal Science. https://doi.org/10.1101/2020.07.10.197426
| A modelling framework for the prediction of the herd-level probability of infection from longitudinal data | Aurélien Madouasse, Mathilde Mercat, Annika van Roon, David Graham, Maria Guelbenzu, Inge Santman Berends, Gerdien van Schaik, Mirjam Nielen, Jenny Frössling, Estelle Ågren, Roger Humphry, Jude Eze, George Gunn, Madeleine Henry, Jörn Gethmann, Sim... | <p>The collective control programmes (CPs) that exist for many infectious diseases of farm animals rely on the application of diagnostic testing at regular time intervals for the identification of infected animals or herds. The diversity of these ... |  | TEST, Veterinary epidemiology | Rowland Raymond Kao | 2020-07-23 08:13:18 | View |
MANAGING BOARD
Karol B Barragán-Fonseca
Mohammed Gagaoua
Rachel Gervais
Florence Gondret
Francois Meurens
Rafael Muñoz-Tamayo
Anna Olsson
Seyed Abbas Rafat
Yuliaxis Ramayo-Caldas









